Genes, proteins and their annotations 
One aspect of the Gentrepid tool is the annotation and gene search engine. As the method of predicting disease gene candidates depends on the protein annotations, we have annotated genes and their products through a number of available analytical and predictive bioinformatic tools, along with other databases and ontologies. These annotations are summarised in the individual gene pages.
Gene pages are compilations of in-house bioinformatic analyses of gene products, with protein isoforms displayed under individual tabs.
Protein domains, coils, and helices are all displayed as an image, with links to the text-based results
of Pfam, SignalP, Marcoil, Multicoil and TMHMM. The mRNA sequences in FASTA format can be accessed, and if applicable, current protein isoform sequences.
Gene pages link to relevant entries in Entrez Gene and GeneCards webservers and the UCSC Genome Browser.
Protein pathways and interactions
Protein pathway data has been retrieved from multiple databases and online resources.
BioCarta and KEGG contain information on signalling and metabolic pathways
respectively.
Protein-protein interaction data are gathered from the Interlogous Interaction
Database (I2D) which contains literature-derived data from BIND, MINT and HPRD
amongst others.

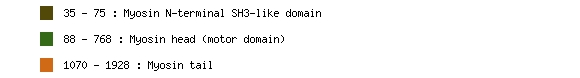
Pfam
Pfam is a large collection of multiple sequence alignments and hidden Markov models covering many common protein domains and families. Pfam can be used to view the domain organisation of proteins, and a protein can have multiple Pfam families. The example below is the domain profile for MYH6, protein NP_002462.


MARCOIL
A hidden Markov model-based program that predicts the existence and location
of potential coiled-coil domains in protein sequences. The prediction program
predicts potential coiled-coil domains at 5 thresholds; 2, 10, 50, 90, 99.
The example below shows the Marcoil prediction for the gene product of SEPHS1,
protein NP_036379.

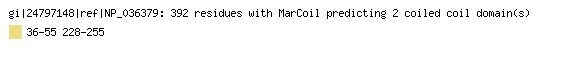

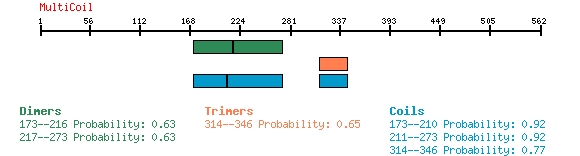
MULTICOIL
Multicoil is used to identify coiled coil regions within a protein sequence,
primarily in the analysis for the identification of trimeric coiled coil
regions (although dimeric and generic coiled coil domains are also reported).
Below is an example of Multicoil predictions for CCDC155 or FLJ32658, protein NP_653289.

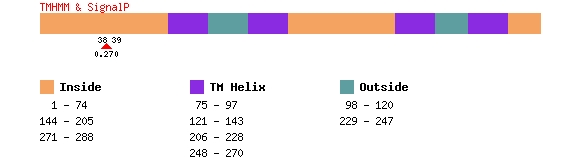
SignalP
Predicts, with a probability, the presence and location of signal peptide
cleavage sites in the first 70 amino acids of the FASTA sequence.

TMHMM
Predicts the presence and location of transmembrane helices in a protein
sequence. This program predicts the regions that are of transmembrane
helix,inside of a cell, or outside of a cell within a protein.
Below is an example of the output for PTPLA, protein NP_055056.

Phenotypes
Phenotype information has been extracted from OMIM.

| 




